Volume 11, Issue 4 (12-2023)
Jorjani Biomed J 2023, 11(4): 11-18 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Abbasi F, Mortazavi N, Behnampour N, Mohammadi M, Mohammadi S. Serum and salivary microRNA-31 in early detection of head and neck squamous cell carcinoma: A systematic review and meta-analysis. Jorjani Biomed J 2023; 11 (4) :11-18
URL: http://goums.ac.ir/jorjanijournal/article-1-996-en.html
URL: http://goums.ac.ir/jorjanijournal/article-1-996-en.html
1- Dental Research Center, Golestan University of Medical Sciences, Gorgan, Iran
2- Dental Research Center, Golestan University of Medical Sciences, Gorgan, Iran. Department of Oral and Maxillofacial Medicine, School of Dentistry, Golestan University of Medical Sciences, Gorgan, Iran ,mortazavi_nazanin@yahoo.com
3- Health Management and Social Development Research Center, Golestan University of Medical Sciences, Gorgan, Iran
4- Medical Librarianship and Information Sciences, Research Center of Gastroenterology and Hepatology, Golestan University of Medical Sciences, Gorgan, Iran
5- Stem Cell Research Center, Golestan University of Medical Sciences, Gorgan, Iran
2- Dental Research Center, Golestan University of Medical Sciences, Gorgan, Iran. Department of Oral and Maxillofacial Medicine, School of Dentistry, Golestan University of Medical Sciences, Gorgan, Iran ,
3- Health Management and Social Development Research Center, Golestan University of Medical Sciences, Gorgan, Iran
4- Medical Librarianship and Information Sciences, Research Center of Gastroenterology and Hepatology, Golestan University of Medical Sciences, Gorgan, Iran
5- Stem Cell Research Center, Golestan University of Medical Sciences, Gorgan, Iran
Full-Text [PDF 688 kb]
(2266 Downloads)
| Abstract (HTML) (7534 Views)
3.2. Exclusion criteria
The lack of access to some scientific databases, the unavailability of the full text of a study, the lack of required results in the studies, and publication biases are the limitations of this study.
4. Statistical analysis
1. Using a funnel plot to detect the distribution bias of primary studies before and after sensitivity analysis
2. Using Cochran's Q test to determine the degree of heterogeneity in the results of primary studies
3. Due to the confirmation of heterogeneity between studies, the model with random effects was used.
4. As the necessary assumptions were established, a cumulative meta-analysis was used.
5. The meta-regression method was also used to determine the cause of heterogeneity, and Stata v. 12 software was used for data analysis.
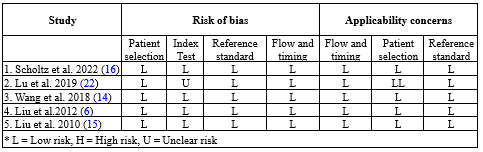
5. Quality assessment of individual studies
The quality assessment of the studies was scored based on the criteria of the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) checklist. The results of the risk of bias assessment are shown in detail in Table 3. For each analyzed category, a judgment with titles of low risk, high risk, or unclear risk was expressed in a report after evaluation by 2 authors. Studies with low risk were considered as good quality studies.

Results
3.1. Systematic review results
In this study, a search was conducted in PubMed, Scopus, Embase, Web of Science, ProQuest, Wiley, and Central Cochrane databases using the described search strategy (Figure 1).
In reviewing the articles, it can be concluded that in patients with HNSCC, miRNA expression may either be elevated or decreased. The miR-21, miR-31, miR-24, miR-494, and miR-155 levels increased, while miR-200, miR-125, and miR-146 levels decreased.
A total of 245 miRNAs were investigated in these studies, of which 8 types of miRNAs were repeated in more than 4 articles. Investigations showed that miR-21, miR-31, and miR-200 had the most repetitions, respectively.
According to Table 4, the investigation of miR-21 in 14 studies showed that this biomarker was prepared by qRT-PCR, TaqMan MiR assays, dSCORE, and NGS from saliva, serum, and plasma samples drawn from the mouth, larynx, and neck regions. MiR-21 increased in 12 articles, and in the articles of Ishinaga (2019) (8) with a sample size of 115 (86 sick people and 29 healthy people) and Yap (2019) (9) with a sample size of 116 (53 sick people and 63 healthy people), a decrease in the amount of this biomarker has been reported.
The investigation of miR-31 in 8 studies showed that this biomarker was prepared from saliva, serum, and plasma samples by qRT-PCR, NGS, and ISAR/Cas12a-dmStrip methods and was examined in the mouth and neck areas. In all studies, an increase in the amount of this biomarker has been reported and was evaluated in the present meta-analysis according to the reported values of sensitivity, specificity, and AUC.
The investigation of miR-200 in 6 studies showed that this biomarker was prepared by RT-qPCR and electrochemical methods from saliva, serum, and plasma samples and was examined only in the oral region. In the article of Wiklund (2011) (10), with a sample size of 32 (25 patients and 7 healthy people), an increase in the amount of this biomarker has been reported. A decrease in the level of this biomarker has been reported in other studies.
The investigation of miR-24 in 4 studies showed that this biomarker was prepared from saliva and serum samples by RT-qPCR, miRNA microarray, and dSCORE methods and was investigated only in the oral region. According to 3 articles, this biomarker has increased, and in the article of Yap et al. (2019) (9) with a sample size of 116 (53 sick people and 63 healthy people), a decrease in the amount of this biomarker has been reported.
The investigation of miR-125 in 4 studies showed that this biomarker was prepared by RT-qPCR, NGS, and electrochemical methods and exclusively from saliva samples and was investigated only in the mouth area. A decrease in this biomarker has been reported in all studies.
The investigation of miR-146 in 5 studies showed that this biomarker was prepared by RT-qPCR, PCR-RFLP, and PCR arrays from saliva and blood samples and was investigated in the mouth and neck area. MiR-146 decreased in 3 articles, and in articles of Yilmaz et al. (2020) (11) with a sample size of 142 (42 sick people and 100 healthy people) and Hung et al. (2013) (12) with a sample size of 63 (51 sick people and 12 healthy people), a rise in the amount of this biomarker has been reported.
The investigation of miR-494 in 4 studies showed that this biomarker was analyzed by RT-qPCR and miRNA microarray methods and exclusively from blood samples and only in the oral region. An increase in the level of this biomarker has been reported in all studies.
The investigation of miR-155 in 4 studies showed that this biomarker was prepared by qRT-PCR, Microarrays, and TaqMan MiR assays from saliva and plasma samples and examined in the mouth and neck areas. In 3 articles, the level of the biomarker has been elevated, and in the article of Lerner et al. (2015) (13) with a sample size of 68 (56 patients and 12 healthy people), a decrease in the amount of this biomarker was reported.
3.2. Meta-analysis results
To perform the meta-analysis, we evaluated MiR-31 for its sensitivity, specificity, and AUC values in 5 validated articles (Table 5, 6). In Figure 2, the information of all 5 studies and their weights based on the fixed model for the feature index can be seen. The meta-analysis estimation of the feature index based on a random model is also reported. The highest weight was assigned to the study of Wang (2018) (14), which is equal to 21.88%. The study by Liu (2010) (15) had the lowest weight (18.16%).
As shown in Figure 3, the information of all 5 studies and their weights based on the fixed model for the AUC index can be seen. The meta-analysis estimation of the AUC index based on a random model is also reported. The highest weight was assigned to the study by Wang (2018) (14), which is equal to 27.69%. Scholtz's study (2022) (16) had the lowest weight (16.54%).
3.2.1 Charts related to publication bias
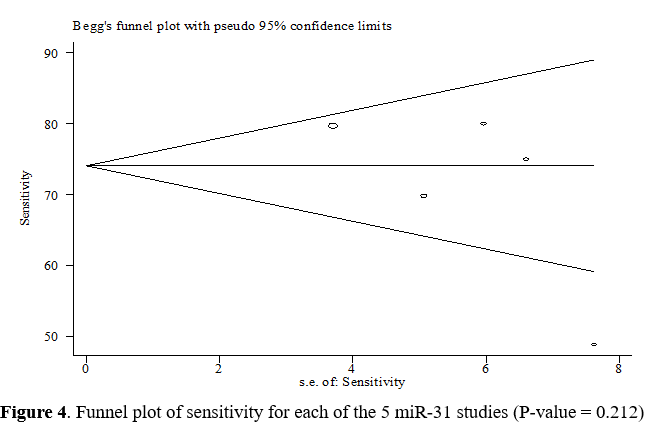
Begg's test and a funnel plot were used to check publication bias. Due to the symmetrical funnel plot in Figure 4, no heterogeneity is apparent between the studies. On the other hand, the P-value for the sensitivity index using Begg's test was equal to 0.212, which indicates the absence of publication bias.
It can be concluded from Figure 5 that there is no heterogeneity between studies since the funnel plot is symmetrical. On the other hand, the P-value for the feature index using Begg's test was equal to 0.325, which indicates the absence of publication bias.
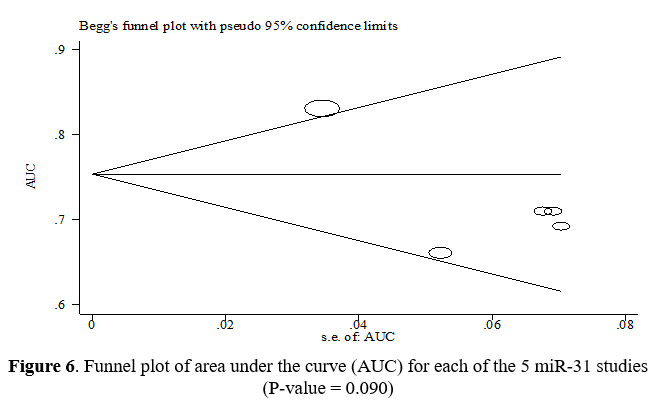
Figure 6 shows that the funnel diagrams are symmetrical, suggesting no heterogeneity between the studies. On the other hand, the P-value for the AUC index using Begg's test was equal to 0.090, which indicates the absence of publication bias.



3.2.2. Sensitivity analysis
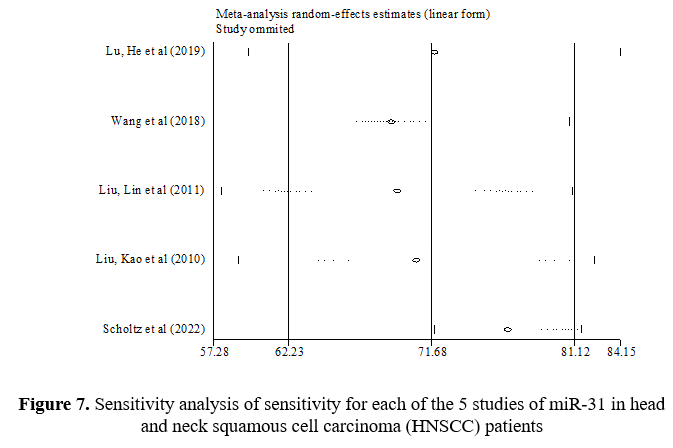
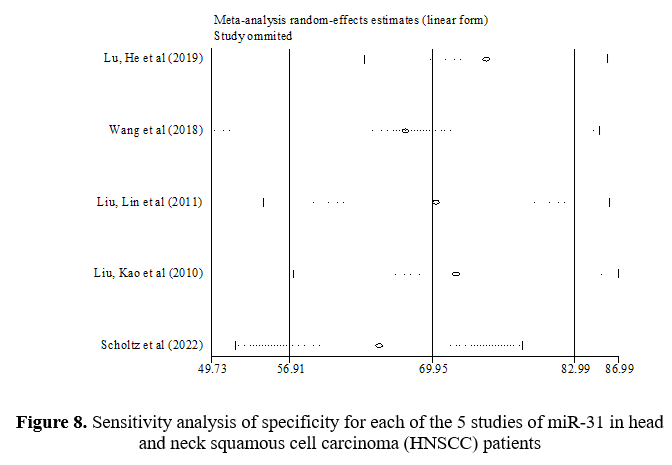
In the sensitivity analysis, the contribution of each study to the overall outcome of the studied index was evaluated. Table 7 shows a sensitivity value of 71.68%, a specificity value of 69.95%, and an AUC value of 0.73. This table shows what the total amount would be if the studies were removed one by one. Figures 7-9 show how each indicator is affected by removing each study.

Discussion
Early detection of oral cancer is essential to have a better prognosis, and studies demonstrated that miRNAs can act as biomarkers for the detection of cancer in its early stage (17).
Specific miRNA expression patterns can not only differentiate cancer cells from normal cells but can also identify the tissue in which the primary tumor originated and can be an early diagnostic marker of HNSCC (18-20).
Our meta-analysis included 5 completely related articles with small amounts of sensitivity, specificity, and AUC in relation to MiR-31. Considering all studies regarding miRNAs, the combined results indicated that AUC = 0.73, with a sensitivity of 71.68% and a specificity of 69.95%, could be used for HNSCC diagnosis. Due to the moderate sensitivity and specificity of miRNAs, they may be able to confirm or exclude suspected cases of this disease, enhancing their utility as clinical diagnostic indicators.
In a meta-analysis by Tian et al., which included 23 studies, combined results indicated that sensitivity, specificity, and AUC were 0.759, 0.773, and 0.832, respectively, indicating a relatively high diagnostic accuracy of miRNAs in differentiating OSCC patients from healthy controls (3).
Another meta-analysis reported that the combined sensitivity and specificity of blood and salivary miRNAs in the diagnosis of OSCC were 0.78 and 0.82, respectively. The overall AUC was 0.91. This study also only included the results of miRNAs in the saliva and blood of patients with OSCC (21).
As a result, miRNAs, especially MiR-31 expression in saliva, serum, or plasma, have the ability to discriminate HNSCC from healthy controls, suggesting that miRNAs are promising molecular markers for the early identification of HNSCC. Since heterogeneity often exists in meta-analyses of diagnostic accuracy data, possible causes that could contribute to the inconsistency in accuracy estimates across the study were investigated, and a random-effect model was used. Interestingly, the funnel plot asymmetry test showed no evidence of publication bias (P-value = 0.325).
Our meta-analysis has a few limitations. First, since all the studies we included were case-control studies, it is unclear how much risk bias might have influenced the quality assessment. Second, when combining the findings of the included articles, we might have overlooked other factors such as different patient stages, HNSCC subtype, HPV (human papillomavirus) status, miRNA normalization methods, and cut-offs.
Therefore, additional studies are needed to compare the diagnostic effect of multiple miRNAs and individual miRNAs on HNSCC. The precise role of miRNAs in the carcinogenesis of tumors remains unknown despite the excellent diagnostic performance achieved by miRNAs in most studies.
Conclusion
The available data provide evidence that miRNAs, especially MiR-31 expression in saliva, serum, or plasma, can be used as a diagnostic biomarker for HNSCC patients. However, controlled clinical trials with large samples are needed to validate different miRNAs.
Acknowledgement
This study was derived from a doctoral dissertation.
Funding sources
This project was funded by Golestan University of Medical Sciences (Grant No. 111791).
Ethical statement
This research was conducted according to ethical guidelines approved by the Ethics Committee of the Golestan University of Medical Sciences (Ethics approval code: IR.GOUMS.REC.1399.368).
Conflicts of interest
The authors declare that there is no conflict of interest.
Author contributions
The authors confirm contribution to the paper as follows: Study conception and design: MN, BN, MM, MS, AF; Bibliographic search and data extraction: MN, AF, MM; Analysis and interpretation of results: BN, MN, AF; Manuscript preparation: AF, MN, BN. All the authors have revised and approved the final version of the manuscript.
Full-Text: (1287 Views)
Introduction
Among all cancers, oral cancer is the sixth most common cancer in the world, coming in third among developing countries (1). The most common type of oral cancer is squamous cell carcinoma (OSCC), which accounts for 90% of all oral cancers and is estimated to affect more than 500,000 people annually (2, 3).
According to recent statistics on cancer deaths among people, early detection is one of the most effective ways to reduce mortality (3). If OSCC is detected in the early stages of cancer (T1), survival chances increase to 80%, and when it is detected later in the cancer course (T3-4), survival rates drop to 20-30% (4).
The diagnostic method of oral cancer is basically based on histopathology (5). Early diagnosis can be made in different ways such as imaging and optical technologies, staining living tissues with toluidine blue, oral preparation of cytology with the help of computer from brush biopsy samples, saliva samples, molecular analysis, and biomarkers including mRNA, miRNA, and proteins (5).
MicroRNAs are short, non-coding RNAs that regulate the translation and degradation of mRNAs and mediate post-transcriptional gene expression. Aberrant microRNA can disrupt normal cell growth cycles and cause malignancy. In general, microRNAs act either as oncogenes or as tumor suppressors in cancers, including oral cancer (6).
Many articles related to microRNA in saliva, serum, and plasma have been reviewed in different types of cancers, including OSCC, stating that this biomarker can be used as a non-invasive method in the diagnosis and prognosis of human cancers (7).
The aim of the present study was to combine the quantitative results of various studies concerning serum and salivary microRNAs for early diagnosis of head and neck squamous cell carcinoma (HNSCC) using the meta-analysis method.
Methods
1. Search strategy and study selection
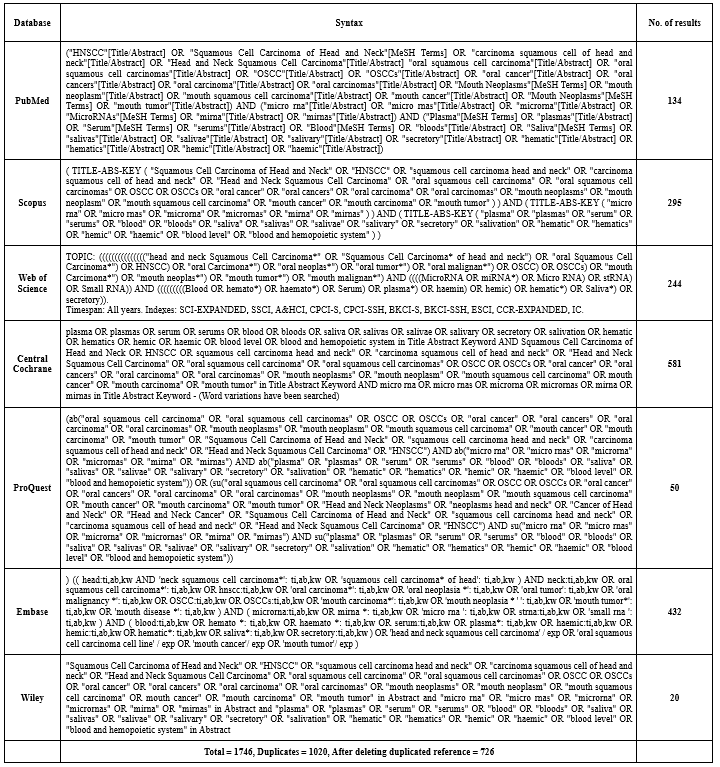
A literature search was conducted for English language case-control articles that investigated various types of miRNAs in serum and saliva for the diagnosis of HNSCC during the years 2000 to 2022, which were indexed and eligible for meta-analysis in databases such as PubMed, Scopus, Web of Science, Central Cochrane, ProQuest, Embase, and Wiley (Table 1). The articles were scored based on the criteria of the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) checklist and the results of risk of bias assessment were determined. For each analyzed category, a judgment with titles of low risk, high risk, or unclear risk was expressed in a report after evaluation by 2 authors.
2. Protocol and registration
The present study is based on the PRISMA checklist (Preferred Reporting Items for Systematic Reviews and Meta-analyses).
Based on a systematic search of databases and registered articles, all eligible articles were identified, and the desired values were extracted (Figure 1). Searching in the titles and abstracts of the studies was done using logical combinations of keywords. Geographical limitations and language limitations were not considered.
The present study has also been registered in the PROSPERO database for possible protocols (CRD42021227449 reference No.).
3. Selection criteria
3.1. Inclusion criteria
Among all cancers, oral cancer is the sixth most common cancer in the world, coming in third among developing countries (1). The most common type of oral cancer is squamous cell carcinoma (OSCC), which accounts for 90% of all oral cancers and is estimated to affect more than 500,000 people annually (2, 3).
According to recent statistics on cancer deaths among people, early detection is one of the most effective ways to reduce mortality (3). If OSCC is detected in the early stages of cancer (T1), survival chances increase to 80%, and when it is detected later in the cancer course (T3-4), survival rates drop to 20-30% (4).
The diagnostic method of oral cancer is basically based on histopathology (5). Early diagnosis can be made in different ways such as imaging and optical technologies, staining living tissues with toluidine blue, oral preparation of cytology with the help of computer from brush biopsy samples, saliva samples, molecular analysis, and biomarkers including mRNA, miRNA, and proteins (5).
MicroRNAs are short, non-coding RNAs that regulate the translation and degradation of mRNAs and mediate post-transcriptional gene expression. Aberrant microRNA can disrupt normal cell growth cycles and cause malignancy. In general, microRNAs act either as oncogenes or as tumor suppressors in cancers, including oral cancer (6).
Many articles related to microRNA in saliva, serum, and plasma have been reviewed in different types of cancers, including OSCC, stating that this biomarker can be used as a non-invasive method in the diagnosis and prognosis of human cancers (7).
The aim of the present study was to combine the quantitative results of various studies concerning serum and salivary microRNAs for early diagnosis of head and neck squamous cell carcinoma (HNSCC) using the meta-analysis method.
Methods
1. Search strategy and study selection
A literature search was conducted for English language case-control articles that investigated various types of miRNAs in serum and saliva for the diagnosis of HNSCC during the years 2000 to 2022, which were indexed and eligible for meta-analysis in databases such as PubMed, Scopus, Web of Science, Central Cochrane, ProQuest, Embase, and Wiley (Table 1). The articles were scored based on the criteria of the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) checklist and the results of risk of bias assessment were determined. For each analyzed category, a judgment with titles of low risk, high risk, or unclear risk was expressed in a report after evaluation by 2 authors.
2. Protocol and registration
The present study is based on the PRISMA checklist (Preferred Reporting Items for Systematic Reviews and Meta-analyses).
Based on a systematic search of databases and registered articles, all eligible articles were identified, and the desired values were extracted (Figure 1). Searching in the titles and abstracts of the studies was done using logical combinations of keywords. Geographical limitations and language limitations were not considered.
The present study has also been registered in the PROSPERO database for possible protocols (CRD42021227449 reference No.).
3. Selection criteria
3.1. Inclusion criteria
- Articles related to the topic
- Case-control articles
- Articles where the samples were from saliva, serum, or plasma
|
Table 1. Syntax used in databases
 |
3.2. Exclusion criteria
- Absence of the full text of the article
- Articles with tissue samples
- Studies other than case-control ones
The lack of access to some scientific databases, the unavailability of the full text of a study, the lack of required results in the studies, and publication biases are the limitations of this study.
4. Statistical analysis
1. Using a funnel plot to detect the distribution bias of primary studies before and after sensitivity analysis
2. Using Cochran's Q test to determine the degree of heterogeneity in the results of primary studies
3. Due to the confirmation of heterogeneity between studies, the model with random effects was used.
4. As the necessary assumptions were established, a cumulative meta-analysis was used.
5. The meta-regression method was also used to determine the cause of heterogeneity, and Stata v. 12 software was used for data analysis.
5. Quality assessment of individual studies
The quality assessment of the studies was scored based on the criteria of the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) checklist. The results of the risk of bias assessment are shown in detail in Table 3. For each analyzed category, a judgment with titles of low risk, high risk, or unclear risk was expressed in a report after evaluation by 2 authors. Studies with low risk were considered as good quality studies.

|
Table 2. Articles divided by databases and reasons for removing them
 |
|
Table 3. Qualitative evaluation of diagnostic studies based on QUADAS-2
 |
Results
3.1. Systematic review results
In this study, a search was conducted in PubMed, Scopus, Embase, Web of Science, ProQuest, Wiley, and Central Cochrane databases using the described search strategy (Figure 1).
In reviewing the articles, it can be concluded that in patients with HNSCC, miRNA expression may either be elevated or decreased. The miR-21, miR-31, miR-24, miR-494, and miR-155 levels increased, while miR-200, miR-125, and miR-146 levels decreased.
A total of 245 miRNAs were investigated in these studies, of which 8 types of miRNAs were repeated in more than 4 articles. Investigations showed that miR-21, miR-31, and miR-200 had the most repetitions, respectively.
According to Table 4, the investigation of miR-21 in 14 studies showed that this biomarker was prepared by qRT-PCR, TaqMan MiR assays, dSCORE, and NGS from saliva, serum, and plasma samples drawn from the mouth, larynx, and neck regions. MiR-21 increased in 12 articles, and in the articles of Ishinaga (2019) (8) with a sample size of 115 (86 sick people and 29 healthy people) and Yap (2019) (9) with a sample size of 116 (53 sick people and 63 healthy people), a decrease in the amount of this biomarker has been reported.
The investigation of miR-31 in 8 studies showed that this biomarker was prepared from saliva, serum, and plasma samples by qRT-PCR, NGS, and ISAR/Cas12a-dmStrip methods and was examined in the mouth and neck areas. In all studies, an increase in the amount of this biomarker has been reported and was evaluated in the present meta-analysis according to the reported values of sensitivity, specificity, and AUC.
The investigation of miR-200 in 6 studies showed that this biomarker was prepared by RT-qPCR and electrochemical methods from saliva, serum, and plasma samples and was examined only in the oral region. In the article of Wiklund (2011) (10), with a sample size of 32 (25 patients and 7 healthy people), an increase in the amount of this biomarker has been reported. A decrease in the level of this biomarker has been reported in other studies.
The investigation of miR-24 in 4 studies showed that this biomarker was prepared from saliva and serum samples by RT-qPCR, miRNA microarray, and dSCORE methods and was investigated only in the oral region. According to 3 articles, this biomarker has increased, and in the article of Yap et al. (2019) (9) with a sample size of 116 (53 sick people and 63 healthy people), a decrease in the amount of this biomarker has been reported.
The investigation of miR-125 in 4 studies showed that this biomarker was prepared by RT-qPCR, NGS, and electrochemical methods and exclusively from saliva samples and was investigated only in the mouth area. A decrease in this biomarker has been reported in all studies.
The investigation of miR-146 in 5 studies showed that this biomarker was prepared by RT-qPCR, PCR-RFLP, and PCR arrays from saliva and blood samples and was investigated in the mouth and neck area. MiR-146 decreased in 3 articles, and in articles of Yilmaz et al. (2020) (11) with a sample size of 142 (42 sick people and 100 healthy people) and Hung et al. (2013) (12) with a sample size of 63 (51 sick people and 12 healthy people), a rise in the amount of this biomarker has been reported.
The investigation of miR-494 in 4 studies showed that this biomarker was analyzed by RT-qPCR and miRNA microarray methods and exclusively from blood samples and only in the oral region. An increase in the level of this biomarker has been reported in all studies.
The investigation of miR-155 in 4 studies showed that this biomarker was prepared by qRT-PCR, Microarrays, and TaqMan MiR assays from saliva and plasma samples and examined in the mouth and neck areas. In 3 articles, the level of the biomarker has been elevated, and in the article of Lerner et al. (2015) (13) with a sample size of 68 (56 patients and 12 healthy people), a decrease in the amount of this biomarker was reported.
3.2. Meta-analysis results
To perform the meta-analysis, we evaluated MiR-31 for its sensitivity, specificity, and AUC values in 5 validated articles (Table 5, 6). In Figure 2, the information of all 5 studies and their weights based on the fixed model for the feature index can be seen. The meta-analysis estimation of the feature index based on a random model is also reported. The highest weight was assigned to the study of Wang (2018) (14), which is equal to 21.88%. The study by Liu (2010) (15) had the lowest weight (18.16%).
As shown in Figure 3, the information of all 5 studies and their weights based on the fixed model for the AUC index can be seen. The meta-analysis estimation of the AUC index based on a random model is also reported. The highest weight was assigned to the study by Wang (2018) (14), which is equal to 27.69%. Scholtz's study (2022) (16) had the lowest weight (16.54%).
3.2.1 Charts related to publication bias
Begg's test and a funnel plot were used to check publication bias. Due to the symmetrical funnel plot in Figure 4, no heterogeneity is apparent between the studies. On the other hand, the P-value for the sensitivity index using Begg's test was equal to 0.212, which indicates the absence of publication bias.
It can be concluded from Figure 5 that there is no heterogeneity between studies since the funnel plot is symmetrical. On the other hand, the P-value for the feature index using Begg's test was equal to 0.325, which indicates the absence of publication bias.
Figure 6 shows that the funnel diagrams are symmetrical, suggesting no heterogeneity between the studies. On the other hand, the P-value for the AUC index using Begg's test was equal to 0.090, which indicates the absence of publication bias.





3.2.2. Sensitivity analysis
In the sensitivity analysis, the contribution of each study to the overall outcome of the studied index was evaluated. Table 7 shows a sensitivity value of 71.68%, a specificity value of 69.95%, and an AUC value of 0.73. This table shows what the total amount would be if the studies were removed one by one. Figures 7-9 show how each indicator is affected by removing each study.



Table 4. Data extracted from final studies with more than 4 microRNA replicates |
Table 5. Data extracted from the articles included in the meta-analysis Table 6. The estimated sensitivity (%), specificity (%), and AUC of mir-31 in preoperative patients and their 95% confidence intervals a  Table 7. Sensitivity analysis of specificity, sensitivity, and AUC for each of 5 studies of miR-31 in head and neck squamous cell carcinoma (HNSCC) patients 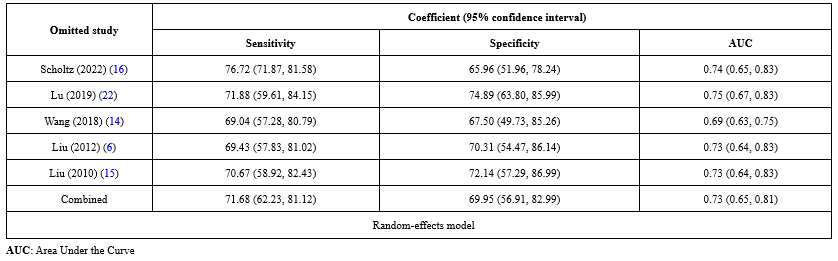 |
Discussion
Early detection of oral cancer is essential to have a better prognosis, and studies demonstrated that miRNAs can act as biomarkers for the detection of cancer in its early stage (17).
Specific miRNA expression patterns can not only differentiate cancer cells from normal cells but can also identify the tissue in which the primary tumor originated and can be an early diagnostic marker of HNSCC (18-20).
Our meta-analysis included 5 completely related articles with small amounts of sensitivity, specificity, and AUC in relation to MiR-31. Considering all studies regarding miRNAs, the combined results indicated that AUC = 0.73, with a sensitivity of 71.68% and a specificity of 69.95%, could be used for HNSCC diagnosis. Due to the moderate sensitivity and specificity of miRNAs, they may be able to confirm or exclude suspected cases of this disease, enhancing their utility as clinical diagnostic indicators.
In a meta-analysis by Tian et al., which included 23 studies, combined results indicated that sensitivity, specificity, and AUC were 0.759, 0.773, and 0.832, respectively, indicating a relatively high diagnostic accuracy of miRNAs in differentiating OSCC patients from healthy controls (3).
Another meta-analysis reported that the combined sensitivity and specificity of blood and salivary miRNAs in the diagnosis of OSCC were 0.78 and 0.82, respectively. The overall AUC was 0.91. This study also only included the results of miRNAs in the saliva and blood of patients with OSCC (21).
As a result, miRNAs, especially MiR-31 expression in saliva, serum, or plasma, have the ability to discriminate HNSCC from healthy controls, suggesting that miRNAs are promising molecular markers for the early identification of HNSCC. Since heterogeneity often exists in meta-analyses of diagnostic accuracy data, possible causes that could contribute to the inconsistency in accuracy estimates across the study were investigated, and a random-effect model was used. Interestingly, the funnel plot asymmetry test showed no evidence of publication bias (P-value = 0.325).
Our meta-analysis has a few limitations. First, since all the studies we included were case-control studies, it is unclear how much risk bias might have influenced the quality assessment. Second, when combining the findings of the included articles, we might have overlooked other factors such as different patient stages, HNSCC subtype, HPV (human papillomavirus) status, miRNA normalization methods, and cut-offs.
Therefore, additional studies are needed to compare the diagnostic effect of multiple miRNAs and individual miRNAs on HNSCC. The precise role of miRNAs in the carcinogenesis of tumors remains unknown despite the excellent diagnostic performance achieved by miRNAs in most studies.
Conclusion
The available data provide evidence that miRNAs, especially MiR-31 expression in saliva, serum, or plasma, can be used as a diagnostic biomarker for HNSCC patients. However, controlled clinical trials with large samples are needed to validate different miRNAs.
Acknowledgement
This study was derived from a doctoral dissertation.
Funding sources
This project was funded by Golestan University of Medical Sciences (Grant No. 111791).
Ethical statement
This research was conducted according to ethical guidelines approved by the Ethics Committee of the Golestan University of Medical Sciences (Ethics approval code: IR.GOUMS.REC.1399.368).
Conflicts of interest
The authors declare that there is no conflict of interest.
Author contributions
The authors confirm contribution to the paper as follows: Study conception and design: MN, BN, MM, MS, AF; Bibliographic search and data extraction: MN, AF, MM; Analysis and interpretation of results: BN, MN, AF; Manuscript preparation: AF, MN, BN. All the authors have revised and approved the final version of the manuscript.
Type of Article: Systematic Review and Meta Analysis |
Subject:
Basic Medical Sciences
Received: 2023/10/28 | Accepted: 2023/12/16 | Published: 2023/12/27
Received: 2023/10/28 | Accepted: 2023/12/16 | Published: 2023/12/27
References
1. Jiang J-S, Zhu J, Han H, Wang Q. Expression of Oct-4 in oncogenic miR-155-positive oral squamous carcinoma cells. 2016;15(9):1847-52. [View at Publisher] [DOI] [Google Scholar]
2. Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, et al. Salivary microRNA: Discovery, Characterization, and Clinical Utility for Oral Cancer Detection. Clin Cancer Res. 2009;15(17):5473-7. [View at Publisher] [DOI] [PMID] [Google Scholar]
3. Tian X, Chen Z, Shi S, Wang X, Wang W, Li N, et al. Clinical diagnostic implications of body fluid miRNA in oral squamous cell carcinoma: A meta-analysis. Medicine (Baltimore). 2015;94(37):e1324. [View at Publisher] [DOI] [PMID] [Google Scholar]
4. Dumache R, Rogobete AF, Andreescu N, Puiu MJCL. Genetic and epigenetic biomarkers of molecular alterations in oral carcinogenesis. 2015;61(10):1373-81. [View at Publisher] [DOI] [Google Scholar]
5. Glick M, William MJS. CT: PMPH-USA Ltd. Burket's Oral medicine. 12th ed. New York: Buffalo; 2015. p.250 [View at Publisher] [Google Scholar]
6. Liu CJ, Lin SC, Yang CC, Cheng HW, Chang KW. neck. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. 2012;34(2):219-24. [View at Publisher] [DOI] [PMID] [Google Scholar]
7. Karimi A, Bahrami N, Sayedyahossein A, Derakhshan SJJoOP. Medicine. Evaluation of circulating serum 3 types of microRNA as biomarkers of oral squamous cell carcinoma; A pilot study. 2020;49(1):43-8. [View at Publisher] [DOI] [PMID] [Google Scholar]
8. Ishinaga H, He F, Hou B, Shah S, Murata M, Takeuchi K. A longitudinal study on circulating miR-21 as a therapeutic effect marker in head and neck squamous cell carcinoma. Carcinogenesis. 2019;40(9):1070-6. [View at Publisher] [DOI] [PMID] [Google Scholar]
9. Yap T, Seers C, Koo K, Cheng L, Vella L, Hill A, et al. Non-invasive screening of a microRNA-based dysregulation signature in oral cancer and oral potentially malignant disorders. Oral Oncol. 2019;96:113-20. [View at Publisher] [DOI] [PMID] [Google Scholar]
10. Wiklund ED, Gao S, Hulf T, Sibbritt T, Nair S, Costea DE, et al. MicroRNA alterations and associated aberrant DNA methylation patterns across multiple sample types in oral squamous cell carcinoma. PLoS ONE. 2011;6(11):e27840. [View at Publisher] [DOI] [PMID] [Google Scholar]
11. Yılmaz K, Gümüşay Ö, Nursal AF, Karakuş N, Yiğit S. Genetic Variations of miRNAs and the Risk of Oral Squamous Cell Carcinoma: A Case-control Study.Medical Bulletin of Haseki/Haseki Tip Bulteni. 2020;58(3):268-73. [View at Publisher] [DOI] [Google Scholar]
12. Hung P-S, Liu C-J, Chou C-S, Kao S-Y, Yang C-C, Chang K-W, et al. miR-146a enhances the oncogenicity of oral carcinoma by concomitant targeting of the IRAK1, TRAF6 and NUMB genes. PLoS ONE. 2013;8(11):e79926. [View at Publisher] [DOI] [PMID] [Google Scholar]
13. Lerner C, Bochen F, Kulas P, Linxweiler M, Hasenfus A, Heinzelmann J, et al. 149 Influence of miR-146a and miR-155 on cell proliferation and migration in head and neck squamous cell carcinoma. Eur J Cancer. 2015;3(51):14. [View at Publisher] [DOI] [Google Scholar]
14. Wang L-L, Li H-X, Yang Y-Y, Su Y-L, Lian J-S, Li T, et al. MiR-31 is a potential biomarker for diagnosis of head and neck squamous cell carcinoma. Int J Clin Exp Pathol. 2018;11(9):4339-45. [View at Publisher] [Google Scholar]
15. Liu CJ, Kao SY, Tu HF, Tsai MM, Chang KW, Lin SC. Increase of microRNA miR-31 level in plasma could be a potential marker of oral cancer. Oral Dis. 2010;16(4):360-4. [View at Publisher] [DOI] [PMID] [Google Scholar]
16. Scholtz B, Horváth J, Tar I, Kiss C, Márton IJJP. Salivary miR-31-5p, miR-345-3p, and miR-424-3p Are Reliable Biomarkers in Patients with. Oral Squamous Cell Carcinoma. 2022;11(2):229. [View at Publisher] [DOI] [PMID] [Google Scholar]
17. Dioguardi M, Caloro GA, Laino L, Alovisi M, Sovereto D, Crincoli V, et al. Circulating miR-21 as a potential biomarker for the diagnosis of oral cancer: A systematic review with meta-analysis. Cancers. 2020;12(4):936. [View at Publisher] [DOI] [PMID] [Google Scholar]
18. Kozaki K-i, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68(7):2094-105. [View at Publisher] [DOI] [PMID] [Google Scholar]
19. Lajer C, Garnaes E, Friis-Hansen L, Norrild B, Therkildsen M, Glud M, et al. The role of miRNAs in human papilloma virus (HPV)-associated cancers: bridging between HPV-related head and neck cancer and cervical cancer. Br J Cancer. 2012;106(9):1526-34. [View at Publisher] [DOI] [PMID] [Google Scholar]
20. Wong TS, Liu XB, Wong BYH, Ng RWM, Yuen APW, Wei WI. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588-92. [View at Publisher] [DOI] [PMID] [Google Scholar]
21. Rapado-González O, Martínez-Reglero C, Salgado-Barreira A, López-López R, Suárez-Cunqueiro M, Muinelo-Romay L. miRNAs in liquid biopsy for oral squamous cell carcinoma diagnosis: Systematic review and meta-analysis. Oral Oncol. 2019;99:104465. [view at publisher] [DOI] [PMID] [Google Scholar]
22. Lu Z, He Q, Liang J, Li W, Su Q, Chen Z, et al. miR-31-5p is a potential circulating biomarker and therapeutic target for oral cancer. Mol Therapy-Nucleic Acids. 2019;16:471-80. [view at publisher] [DOI] [PMID] [Google Scholar]
23. Mahmood N, Hanif M, Ahmed A, Jamal Q, Mushtaq S, Khan A, et al. Circulating miR-21 as a prognostic and predictive biomarker in oral squamous cell carcinoma. Pak J Med Sci. 2019;35(5):1408-12. [view at publisher] [DOI] [PMID] [Google Scholar]
24. Mehdipour M, Shahidi M, Manifar S, Jafari S, Abbas FM, Barati M, et al. Diagnostic and prognostic relevance of salivary microRNA-21,-125a,-31 and-200a levels in patients with oral lichen planus-a short report. Cell Oncol. 2018;41(3):329-34. [view at publisher] [DOI] [PMID] [Google Scholar]
25. Momen-Heravi F, Bala S. Extracellular vesicles in oral squamous carcinoma carry oncogenic miRNA profile and reprogram monocytes via NF-κB pathway. Oncotarget. 2018;9(78):34838-34854. [view at publisher] [DOI] [PMID] []
26. Singh P, Srivastava AN, Sharma R, Mateen S, Shukla B, Singh A, et al. Circulating microRNA-21 expression as a novel serum biomarker for oral sub-mucous fibrosis and oral squamous cell carcinoma. Asian Pac J Cancer Prev. 2018;19(4):1053-7. [view at publisher] [DOI] [PubMed] [Google Scholar]
27. Yap T, Koo K, Cheng L, Vella LJ, Hill AF, Reynolds E, et al. Predicting the Presence of Oral Squamous Cell Carcinoma Using Commonly Dysregulated MicroRNA in Oral SwirlsOSCC Prediction from MicroRNA in Oral Swirls. 2018;11(8):491-502. [view at publisher] [DOI] [PMID] [Google Scholar]
28. Wei L, Mao M, Liu H. Droplet digital PCR and qRT-PCR to detect circulating miR-21 in laryngeal squamous cell carcinoma and pre-malignant laryngeal lesions. Acta Otolaryngol. 2016;136(9):923-32. [view at publisher] [DOI] [PMID] [Google Scholar]
29. Yan Y, Wang X, Venø MT, Bakholdt V, Sørensen JA, Krogdahl A, et al. Circulating miRNAs as biomarkers for oral squamous cell carcinoma recurrence in operated patients. Oncotarget. 2017;8(5):8206-14. [view at publisher] [DOI] [PMID] []
30. Zahran F, Ghalwash D, Shaker O, Al-Johani K, Scully C. Salivary micro RNA s in oral cancer. Oral Dis. 2015;21(6):739-47. [view at publisher] [DOI] [PMID] [Google Scholar]
31. Ren W, Qiang C, Gao L, Li S-M, Zhang L-M, Wang X-L, et al. Circulating microRNA-21 (MIR-21) and phosphatase and tensin homolog (PTEN) are promising novel biomarkers for detection of oral squamous cell carcinoma. Biomarkers. 2014;19(7):590-6. [view at publisher] [DOI] [PMID] [Google Scholar]
32. Hsu C-M, Lin P-M, Wang Y-M, Chen Z-J, Lin S-F, Yang M-Y. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumor Biology. 2012;33(6):1933-42. [view at publisher] [DOI] [PMID] [Google Scholar]
33. Jung HM, Phillips BL, Patel RS, Cohen DM, Jakymiw A, Kong WW, et al. Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer. J Biol Chem. 2012;287(35):29261-72. [view at publisher] [DOI] [PMID] []
34. Chen M, Luo R, Li S, Li H, Qin Y, Zhou D, et al. based strip for ultrasensitive detection of OSCC-associated salivary microRNA via CRISPR/Cas12a coupling with IS-primer amplification reaction. Anal Chem. 2020;92(19):13336-42. [view at publisher] [DOI] [PMID] [Google Scholar]
35. Yan Y, Yan F, Huang Q. MiR-200c inhibited the proliferation of oral squamous cell carcinoma cells by targeting Akt pathway and its downstream Glut1. Arch Oral Biol. 2018;96:52-7. [view at publisher] [DOI] [PMID] [Google Scholar]
36. Sun G, Cao Y, Wang P, Song H, Bie T, Li M, et al. miR-200b-3p in plasma is a potential diagnostic biomarker in oral squamous cell carcinoma. Biomarkers. 2018;23(2):137-41. [view at publisher] [DOI] [PMID] [Google Scholar]
37. Wang Z, Zhang J, Guo Y, Wu X, Yang W, Xu L, et al. A novel electrically magnetic-controllable electrochemical biosensor for the ultra sensitive and specific detection of attomolar level oral cancer-related microRNA. Biosens Bioelectron. 2013;45:108-13. [view at publisher] [DOI] [PMID] [Google Scholar]
38. He L, Ping F, Fan Z, Zhang C, Deng M, Cheng B, et al. Salivary exosomal miR-24-3p serves as a potential detective biomarker for oral squamous cell carcinoma screening. Biomed Pharmacother. 2020;121:109553. [view at publisher] [DOI] [PMID] [Google Scholar]
39. Lin S-C, Liu C-J, Lin J-A, Chiang W-F, Hung P-S, Chang K-W. miR-24 up-regulation in oral carcinoma: positive association from clinical and in vitro analysis. Oral Oncol. 2010;46(3):204-8. [view at publisher] [DOI] [PMID] [Google Scholar]
40. Salazar-Ruales C, Arguello J-V, López-Cortés A, Cabrera-Andrade A, Garcia-Cardenas JM, Guevara-Ramirez P et al. Salivary MicroRNAs for early detection of head and neck squamous cell carcinoma: A case-control study in the high altitude mestizo ecuadorian population. BioMed research international. 2018;2018. [view at publisher] [DOI] [PMID] []
41. Min S-K, Jung SY, Kang HK, Park S-A, Lee JH, Kim M-J, et al. Functional diversity of miR-146a-5p and TRAF6 in normal and oral cancer cells. Int J Oncol. 2017;51(5):1541-52. [view at publisher] [DOI] [PMID] [Google Scholar]
42. Emami N, Mohamadnia A, Mirzaei M, Bayat M, Mohammadi F, Bahrami N. miR-155, miR-191, and miR-494 as diagnostic biomarkers for oral squamous cell carcinoma and the effects of Avastin on these biomarkers. J Korean Association Oral Maxillofacial Surg. 2020;46(5):341-7. [view at publisher] [DOI] [PMID] []
43. Ries J, Baran C, Wehrhan F, Weber M, Neukam FW, Krautheim-Zenk A, et al. Prognostic significance of altered miRNA expression in whole blood of OSCC patients. Oncol Rep. 2017;37(6):3467-74. [view at publisher] [DOI] [PMID] [Google Scholar]
44. Ries J, Vairaktaris E, Agaimy A, Kintopp R, Baran C, Neukam FW, et al. miR-186, miR-3651 and miR-494: potential biomarkers for oral squamous cell carcinoma extracted from whole blood. Oncol Rep. 2014;31(3):1429-36. [view at publisher] [DOI] [PMID] [Google Scholar]
45. Ries J, Vairaktaris E, Kintopp R, Baran C, Neukam FW, Nkenke E. Alterations in miRNA expression patterns in whole blood of OSCC patients. In vivo. 2014;28(5):851-61. [view at publisher] [PubMed] [Google Scholar]
46. Narimani A, Hosseini F, Bahrami N, Mohamadnia A. The Expression of MicroRNA-155 (miR-155) and Carcinoembryonic Antigen Messenger RNA (CEA mRNA) in Peripheral Blood of Patients with Oral Squamous Cell Carcinomas (OSCC). J Isfahan Med Sch. 2019; 36(510):1597-601. [view at publisher] [DOI] [Google Scholar]
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |






